Data Call
Data call for tree species annotations and aerial RGB or RGBI imagery (TreeAI)
Deadline for submission: ongoing
Looking for data and collaborators
As part of the COST action 3DForEcoTech and COST SNSF project, we aim to establish an international collaboration and create the first database for the detection of individual tree species.
We are seeking:
- manual tree species canopy delineations.
- aerial RGBI (red, green, blue, and near infrared) or RGB imagery data for the delineated tree species from around the world.
The TreeAI database will serve as the foundation for training a universal deep-learning model for monitoring tree species using fine-grained aerial data.
We are also looking for:
- people with experience in deep learning for object detection or instance segmentation.
- people who are willing to contribute to the writing of the manuscripts.
Your benefit:
You will be given the opportunity to contribute as a co-author in
- the publication of a data manuscript (the data provided will only be published with your consent).
- a manuscript investigating different approaches to detecting individual tree species.
Consortium:
- Dr. Mirela Beloiu Schwenke, Zhongyu Xia, Prof. Verena Griess (ETH Zurich)
- Dr. Martin Mokros (University College London)
- Prof. Xinlian Liang (Wuhan University)
- Dr. Stefano Puliti (Norwegian Institute of Bioeconomy Research)
- Dr. Lars Waser, Dr. Natalia Rehush (WSL Switzerland)
- Prof. Teja Kattenborn (University of Freiburg)
- Clemens Mosig (University of Leipzig)
- Yan Cheng (University of Copenhagen)
Contact:
Zhongyu Xia, zhongyu.xia[at]usys.ethz.ch
Dr. Mirela Beloiu Schwenke, mirela.beloiu[at]usys.ethz.ch
Deadline: ongoing
The criteria for data submissions are as follows:
- RGBI (or RGB) imagery (at ≤ 10 cm, about 3.94 in, spatial resolution) of the site.
- Ground truth tree data (n>100), including two minimum attributes:
- Geolocation of individual tree with manual delineation of the individual tree canopy (strongly preferred) AND/OR coordinates of the point of the individual tree canopy.
- Individual tree species records. The data might originate from either forests or urban regions.
Ground truth tree data: For the delineation of the tree crowns a shapefile of the individual trees with the two minimum attributes. All the trees in one image should be delineated (strong preference) (see Fig. 1 as example). Partial delineations are also accepted, but please mention that in the submission form. GPS data with the coordinates of the tree species (no delineation of the tree crown) are only accepted if the tree size or diameter at breast height is specified for each tree in the attribute table. Please make sure that the coordinates are in the WGS 1984 UTM Coordinate Reference System (CRS). If available, a polygon with the study area should be provided.
Latin names for species: Please provide a column in the attribute table named Species with the Latin species names e.g. Picea abies (use capital letter for the genus name, but not for the species name i.e. not Picea Abies L.). In case the species is not known, the genus should be reported as Genus sp. such as in Picea sp. Dead trees should be named: Dead tree.
RGBI images: tiff file. An orthomosaic for the study region and each year is preferred, yet clips of the plots are also acceptable. Please specify the acquisition date. Make sure that the order of the bands is RGBI.
Coordinate system: Please provide images and ground truth in the Projected Coordinate System WGS 1984 UTM (the zone will depend on your geographic location).
COCO format: A JSON annotation file with the attributes and the image files (e.g., png, jpg, or tiff), labels (.txt), and metadata with the explanation of the classes and the data.

We welcome ground truth tree data with more additional attributes, as this would facilitate further evaluations and developments of models in the future. They include, but are not limited to:
- Time of the measurements of ground truth data (multi-temporal data are welcomed).
- Tree size: diameter at breast height (cm), tree height (m), and social status i.e. dominant and co-dominant.
- Forest density: trees per unit area and basal area per unit area.
- Individual tree species defoliation and mortality (percentage 0 = no defoliation, 100 = dead).
- Environmental and/or topographic measurements: climate, soil, elevation, slope, aspect and so forth.
- Stand age, regeneration conditions (i.e., presence or absence, numbers of saplings), and management operations. For the plots with RGB/RGB-NIR images, the inclusion of high-resolution multispectral/hyperspectral images, high-resolution ground panorama photographs, UAV and/or terrestrial LiDAR point clouds data will be highly valuable.
Short explanation of the license meaning:
- Public (CC BY): Maximizes the dataset’s accessibility and usage, including commercial uses, but ensures you are credited.
- Public (CC BY) - no coordinates: The data is provided in Yolo format without coordinates, which is useful for computer vision tasks.
- Public noncommercial (CC BY-NC-ND) - Only noncommercial uses of the work are permitted, but ensures you are credited.
- Private - the data will only be used internally, not passed on or made available for download.
Note: All data is used for model training. We encourage open data; this will be our first paper's basis.
Please refer to this website for a comprehensive explanation on the license types.
Join our competition!
We are organizing a data science competition for tree species identification between March and June 2025. All details are available on this page: TreeAI4Species Competition.
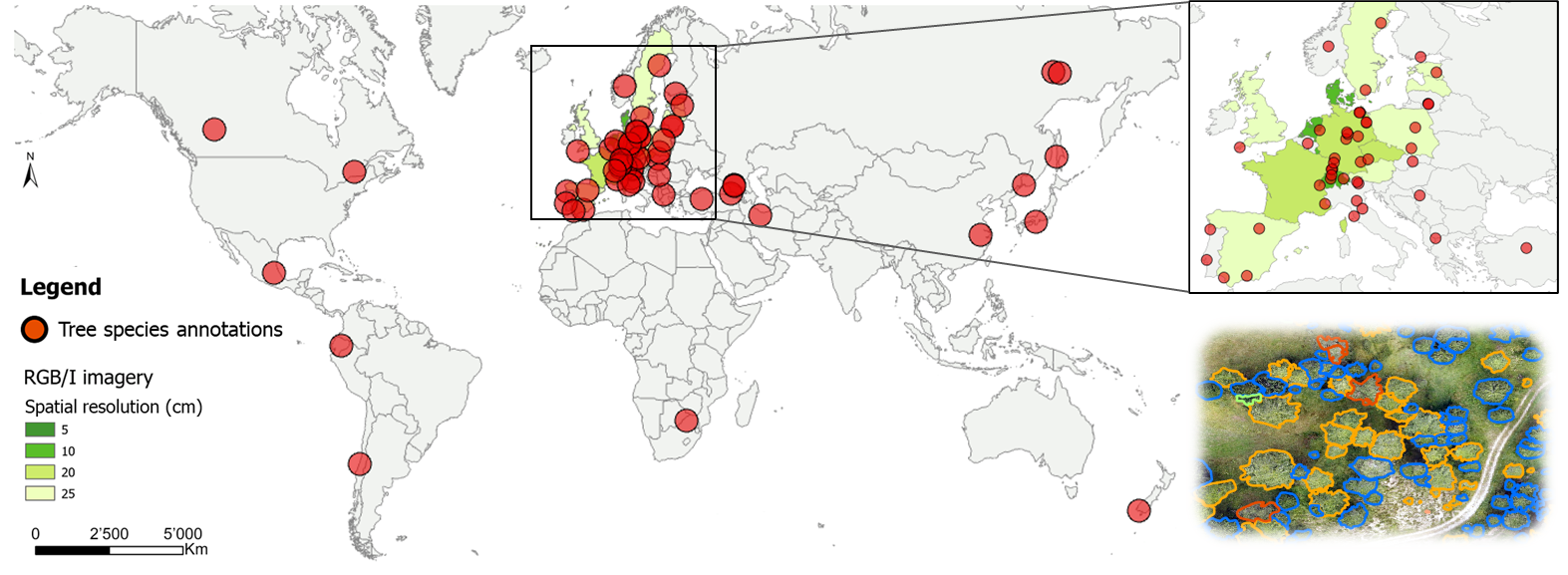
Dataset map
Your dataset might be the next red dot! We currently have RGB/I images and the corresponding tree species annotations from the sites represented by a red dot on this map.
Data Submission Form
Confirmation
Thank you for filling out the form. Now please click on external page this link to upload your file(s) to our Dropbox.